Sanger Sequencing: Principle, Steps, Applications, Diagram

It is also known as dideoxy sequencing or chain termination method. This method was the first DNA sequencing method developed in 1977 by Frederick Sanger and his colleagues. It is considered the gold standard of sequencing due to its high accuracy and ability to produce long reads which is useful for various research applications.
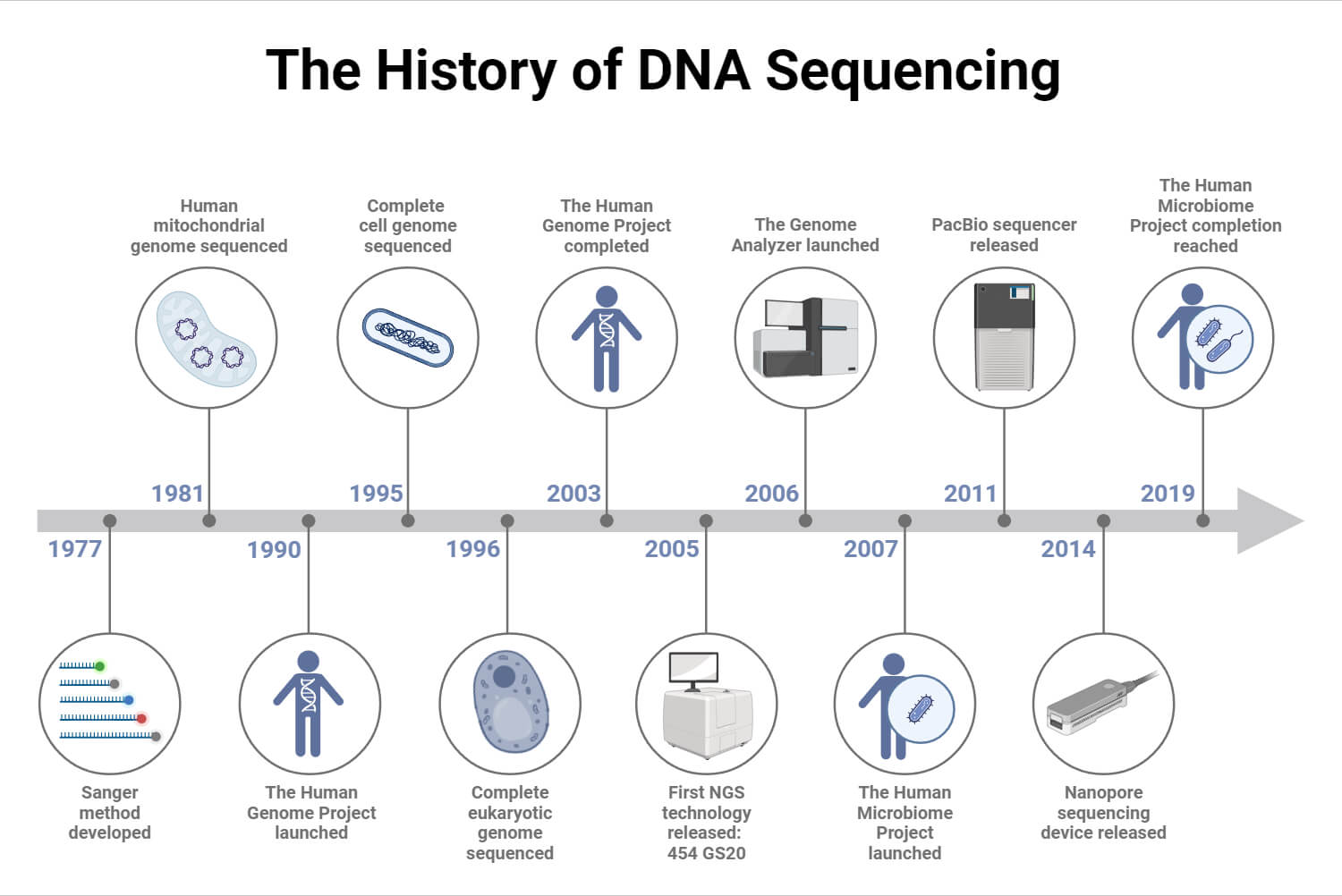
Sequencing technologies have evolved significantly over the past years but Sanger sequencing continues to be the most widely used technology. It is particularly effective for small sequencing projects. However, for higher-throughput sequencing projects, next-generation sequencing (NGS) technologies such as Illumina, PacBio, and Nanopore have emerged. Although NGS technologies have largely replaced Sanger sequencing due to their ability to sequence larger amounts of DNA more quickly and at a lower cost, Sanger sequencing remains in use today. The development of newer DNA sequencing platforms has led to the automation of Sanger sequencing.
Table of Contents
Interesting Science Videos
Principle of Sanger Sequencing
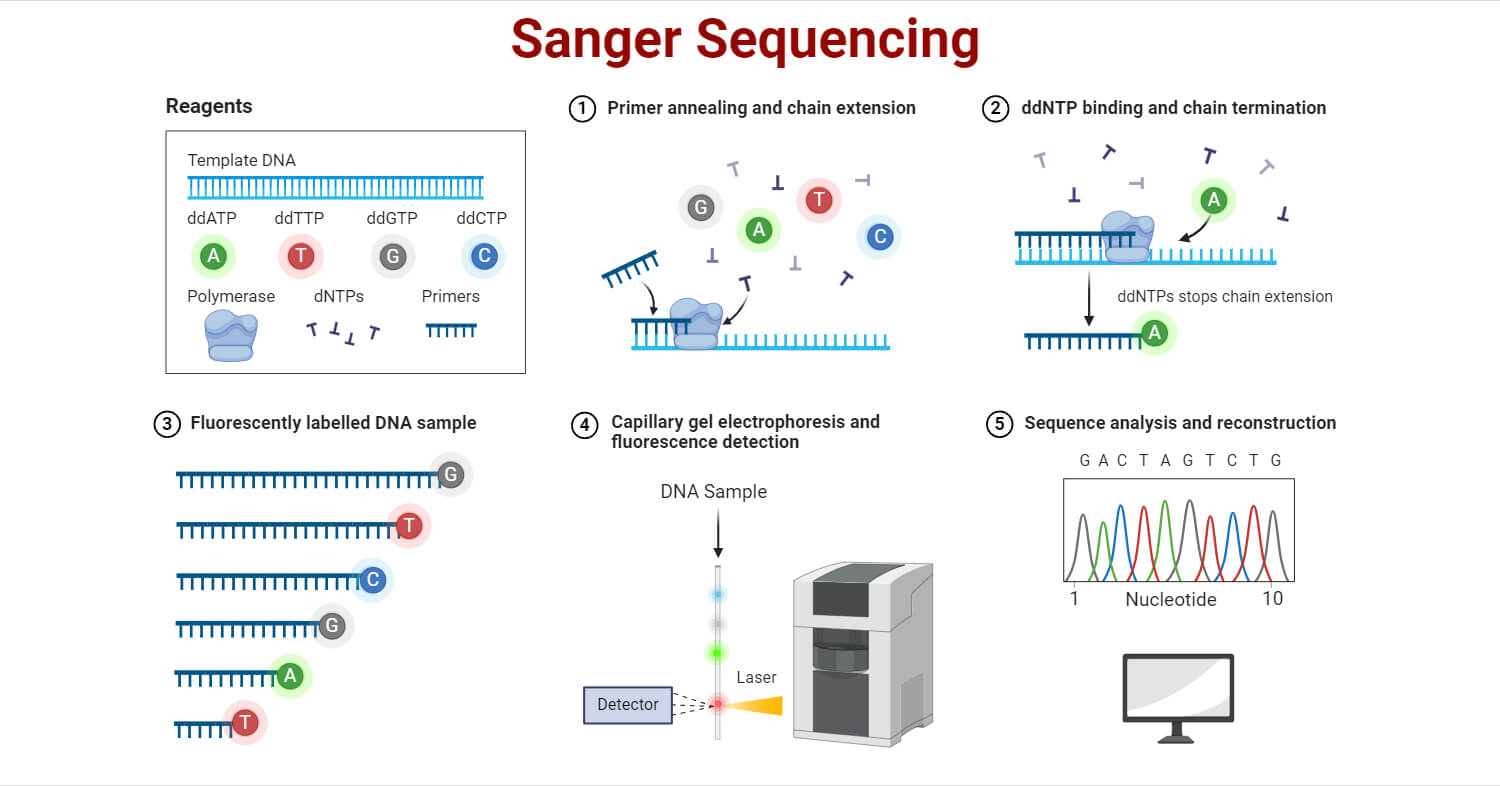
The principle of Sanger sequencing is based on the termination of DNA strand elongation by ddNTPs. These modified molecules are chemical analogs of DNA nucleotides that lack the 3’ hydroxyl group necessary for the formation of a phosphodiester bond that elongates the DNA strand. The addition of ddNTPs in the Polymerase Chain Reaction (PCR) reaction terminates DNA elongation.
During the sequencing process, labeled ddNTPs, deoxyribonucleotide triphosphates (dNTPs), and template DNA are mixed in a PCR reaction. When ddNTPs are added, they terminate the DNA chain, producing fragments of different lengths. These fragments are separated by electrophoresis. The fluorescent labels on the ddNTPs indicate which base terminated each fragment and are used to determine the DNA sequence.
Steps Involved in Sanger Sequencing
Sanger Sequencing involves the following 4 steps.
1. DNA Template Preparation
- The first step in Sanger sequencing is DNA template preparation. At first, the DNA of interest should be extracted from the source.
- There are several methods to extract DNA including chemical extraction, column-based extraction, and magnetic bead-based extraction.
- Identical, single-stranded molecules are prepared for sequencing.
2. Chain Termination PCR
- The target DNA is then amplified using PCR which starts with initial denaturation followed by multiple cycles of denaturation, annealing, and extension and ends with a final hold at 4°C. The PCR reaction includes template DNA, primers, all four dNTPs, and DNA polymerase.
- Along with the dNTPs, a small amount of the four ddNTPs is also added. Each ddNTP is labeled with a distinct fluorescent marker. ddNTPs lack the 3’ hydroxyl group needed for further elongation causing chain termination.
- ddNTPs are present in smaller amounts compared to dNTPs so chain termination occurs at various lengths. This results in a collection of newly synthesized DNA strands of different lengths, each terminating at a ddNTP.
- In the traditional sequencing method, the PCR reaction is performed in four separate tubes, each containing one type of labeled ddNTP.
- In automated Sanger sequencing, all four ddNTPs are included in a single reaction, each labeled with a unique fluorescent marker.
- Sequencing clean-up is done before electrophoresis which removes unincorporated ddNTPs and other contaminants.
3. Separation of DNA Fragments
- Electrophoresis is performed to separate the DNA fragments according to their lengths. The DNA fragments are separated either in polyacrylamide gel or capillary gel system.
- Traditional method separates DNA fragments using polyacrylamide gel electrophoresis which separates the fragments by size. Fragments from each PCR reaction are run in separate lanes to identify the corresponding ddNTP.
- Capillary electrophoresis (CE) uses glass capillaries that are filled with a gel polymer. Each sequencing reaction is run in a single capillary.
4. Detection and Analysis
- After separation, the DNA fragments are passed through a fluorescence detector.
- The detector identifies the fluorescent label attached to the ddNTP at the end of each DNA fragment. Each fluorescent signal represents the nucleotide at the end of the terminated fragment.
- The emitted signals from the nucleotides are captured and a chromatograph is generated, showing the fluorescent peaks of each labeled fragment corresponding to the DNA sequence.
- The sequence of the DNA template is deduced by reading the order of the fluorescent labels corresponding to the terminated DNA fragments.
Sanger Sequencing Steps Video

Advantages of Sanger Sequencing
- Sanger sequencing is considered the gold standard method for many research and clinical applications. It provides highly accurate results. This accuracy is particularly useful for validating sequences obtained through other methods, such as NGS technologies.
- Sanger sequencing technology is well-established with a straightforward process. This leads to reliable and reproducible outcomes.
- Sanger sequencing is suitable for small-scale projects or those including short DNA regions.
- Sanger sequencing produces relatively long read lengths.
- Data analysis from Sanger sequencing does not require complex bioinformatics tools and expertise often necessary for NGS data interpretation.
- The quality of the data obtained from Sanger sequencing is high with clear and easily interpretable chromatograms.
Limitations of Sanger Sequencing
- Sanger sequencing can only sequence short fragments of DNA. It has a limited throughput. It can sequence only one fragment at a time, making it slow and expensive for large genomes.
- Sanger sequencing is not suitable for long DNA sequences or large-scale sequencing projects due to its complexity and cost.
- It involves manual steps compared to NGS workflows and is a time-consuming method. The preparation and handling time for Sanger sequencing can be longer.
- It has low sensitivity which makes it hard to detect rare mutations and study complex mixtures.
- Sanger sequencing requires relatively high-quality and pure DNA samples as degraded or contaminated DNA can lead to unreliable results.
Applications of Sanger Sequencing
- Sanger sequencing is used in medical diagnosis to identify genes associated with different diseases. It is useful for targeted sequencing of specific regions of the genome making it a useful tool in clinical diagnostics and genetic testing.
- Sanger sequencing can be used to identify new species by comparing them with gene sequences of already known species. It is also useful to study the evolutionary history of many species.
- In forensic science, sanger sequencing is useful in personal identification and DNA fingerprinting. This helps us analyze DNA evidence in criminal cases.
- Sanger sequencing is also useful in agriculture to identify different breeds of crops and livestock. It can be used in the conservation of these species.
- Sanger sequencing also finds applications in newer technologies including single-cell sequencing and synthetic biology.
Sanger Sequencing vs. Next-Generation Sequencing
| Characteristics | Sanger sequencing | Next-generation sequencing |
| Principle | It uses the chain termination method and is hence also called dideoxy chain termination sequencing. | It uses parallel sequencing technologies. It is also known as massively parallel sequencing. |
| Speed | This method is comparatively slower. It can process only one fragment at a time. | NGS technologies are fast. It can sequence millions of fragments simultaneously. |
| Read length | It usually produces long read lengths. | Read lengths vary based on the platform but they usually generate short reads. |
| Cost | The cost is high for large-scale projects and low for single genes. | It is economical for high-throughput projects. |
| Data analysis | Data analysis is straightforward. | It requires complex bioinformatics and analysis tools. |
| Applications | This technology is used in small-scale projects and to confirm NGS data. | It is used in large-scale projects and whole-genome sequencing. |
References
- Brown, T. (2010). Gene Cloning and DNA Analysis: An Introduction. A John Wiley & Sons, Ltd., Publication.
- Crossley, B. M., Bai, J., Glaser, A., Maes, R., Porter, E., Killian, M. L., Clement, T., & Toohey-Kurth, K. (2020). Guidelines for Sanger sequencing and molecular assay monitoring. Journal of Veterinary Diagnostic Investigation: official publication of the American Association of Veterinary Laboratory Diagnosticians, Inc, 32(6), 767–775. https://doi.org/10.1177/1040638720905833
- Heather, J. M., & Chain, B. (2016). The sequence of sequencers: The history of sequencing DNA. Genomics, 107(1), 1–8. https://doi.org/10.1016/j.ygeno.2015.11.003
- How to Conduct Sanger Sequencing | Thermo Fisher Scientific – NP
- Sanger Sequencing Steps & Method (sigmaaldrich.com)
- Sanger Sequencing: Introduction, Workflow, and Applications – CD Genomics (cd-genomics.com)
- Slatko, B. E., Gardner, A. F., & Ausubel, F. M. (2018). Overview of Next-Generation Sequencing Technologies. Current protocols in molecular biology, 122(1), e59. https://doi.org/10.1002/cpmb.59
- What is Sanger sequencing? | Thermo Fisher Scientific – NP
About Author
Sanju Tamang completed her Bachelor's (B.Tech) in Biotechnology from Kantipur Valley College, Lalitpur, Nepal. She is interested in genetics, microbiome, and their roles in human health. She is keen to learn more about biological technologies that improve human health and quality of life.





